Sea star killer unmasked. Next step recovery.
The culprit behind the largest known marine disease outbreak is now known, giving researchers new tools to protect and reintroduce these keystone predators.
By Warren Cornwall in Anthropocene August 13, 2025
For the last dozen years, scientists have been on the hunt for a killer that has claimed billions of lives. They’ve finally found it.
In 2013, researchers from Olympic National Park reported what looked like a sea star massacre: ochre sea stars with limbs that had split off from their decaying bodies. It was the first of what soon became a coast-wide underwater epidemic stretching from Mexico to Alaska.
Within years, the mysterious condition, dubbed sea star wasting disease, had wiped out billions of sea stars. It was declared the largest known disease outbreak in the open ocean. The effects were both devastating and gruesome for more than 20 species. Sea stars broke apart, their arms crawling away seemingly in a failed attempt to escape before dissolving into goo.
Many-armed sunflower sea stars as big as bicycle wheels were some of the hardest hit, declining by 99% in U.S. coastal waters and earning the designation of “critically endangered” from the International Union for Conservation of Nature.
Scientists struggled to figure out what was behind this devastation. Initial suspicions of a kind of virus proved wrong. Warming waters appeared to play a role, but that in itself couldn’t explain it.
Starting in 2021, Canadian and U.S. scientists mounted a massive, 4-year hunt to find the culprit. Last week, they announced the results in Nature Ecology & Evolution: a bacteria called Vibrio pectenicida, part of a family of particularly nasty pathogens known to cause everything from cholera to scallop-killing outbreaks.
The discovery is a critical first step in figuring out how to protect or restore sea stars, which are linchpins of many coastal ecosystems such as kelp forests. Those forests are in decline partly because they are being devoured by sea urchins, once prey to sea stars. “Now that we’ve identified the disease-causing agent, we can start looking at how to mitigate the impacts of this epidemic,” said Melanie Prentice, a scientist at the University of British Columbia involved in the research.
The sleuthing involved years spent painstakingly narrowing down the possible causes of the disease, much of it at a U.S. Geological Survey laboratory in Washington state equipped to handle waterborne diseases.
The remarkable restoration of a degraded coastline brought on by returning sea otters
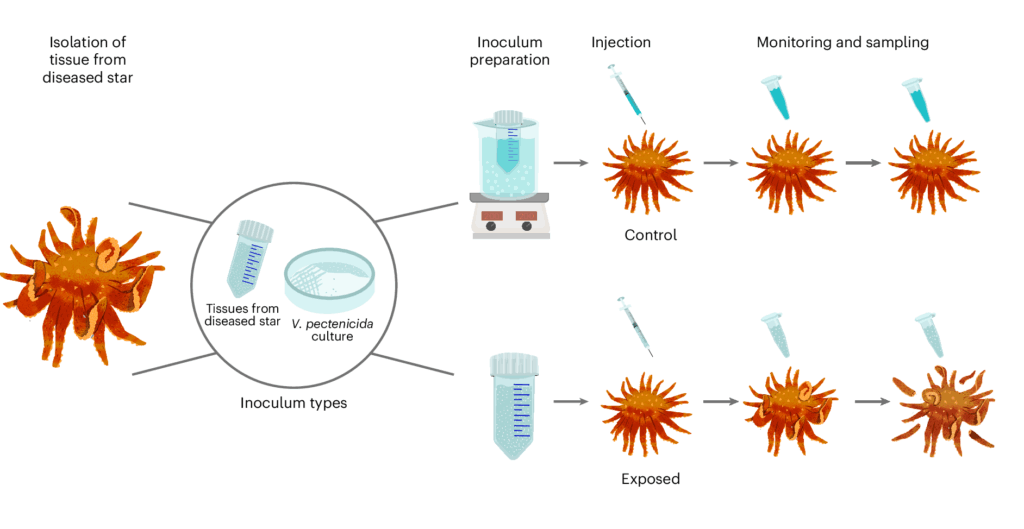
First, scientists tried different ways of exposing healthy sea stars: they put them in tanks with infected ones; added water from tanks with sick sea stars; and injected the sea stars with tissue from infected ones. All approaches proved deadly. Of 50 healthy sea stars, 46 succumbed.
The researchers zeroed in on a substance called coelomic fluid, likened to sea star blood. When sea stars were injected with the fluid from an infected individual, they grew sick. But when they received a version that had been heat-treated to kill live organisms, they remained healthy.
When the DNA of the contents of coelemic fluid from healthy and sick sea stars was scrutinized, the sick ones contained a lot of DNA from the Vibrio bacteria.
“When we looked at the coelomic fluid between exposed and healthy sea stars, there was basically one thing different: Vibrio,” said Alyssa Gehman, a marine disease ecologist at the Hakai Institute and the University of British Columbia. “We all had chills. We thought, ‘That’s it. We have it. That’s what causes wasting.’”
As a final test, they refined a pure sample of the bacteria, then injected it into 6 sunflower sea stars, while another 6 received doses inactivated by high heat. The ones with the live bacteria all died, while the others all survived.
“This is the discovery of the decade for me,” said Drew Harvell, an ecologist with the University of Washington and author of several books about ocean life “What’s crazy is that the answer was just sitting right there in front of us. This Vibrio is a sneaky critter because it doesn’t show up on histology like other bacteria do.”
Other factors, such as heat, might still play a role. It’s not known how the disease first reached sea stars on this coast. But Vibrio bacteria generally thrive in warmer conditions. In fact, scientists have called them a “barometer of climate change.”
The new discovery doesn’t mean scientists will be able to find a “cure.” But it can help guide their work to find sea stars that are resistant to the disease. And researchers can now monitor for outbreaks in the wild by taking water or tissue samples. That might help them decide where to release lab-raised sea stars to give them the best chance of surviving.
“This finding opens up exciting avenues to expand the network of researchers able to develop solutions for recovery of the species,” said Jono Wilson, head of ocean science for The Nature Conservancy’s California chapter, which helped fund the research. “We are actively pursuing studies looking at genetic associations with disease resistance, captive breeding and experimental introduction of captively-raised stars back into the wild.”
Prentice, et. al. “Vibrio pectenicida Vibrio pectenicida strain FHCF-3 is a causative agent of sea star wasting disease.” Nature Ecology & Evolution. Aug. 4, 2025.
References
- Eisenlord, M. E. et al. Ochre star mortality during the 2014 wasting disease epizootic: role of population size structure and temperature. Philos. Trans. R. Soc. B 371, 20150212 (2016).Google Scholar
- Hewson, I. et al. Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl Acad. Sci. USA 111, 17278–17283 (2014).CAS PubMed PubMed Central Google Scholar
- Dawson, M. N. et al. A decade of death and other dynamics: deepening perspectives on the diversity and distribution of sea stars and wasting. Biol. Bull. 244, 143–163 (2023).PubMed Google Scholar
- Harvell, C. D. et al. Disease epidemic and a marine heat wave are associated with the continental-scale collapse of a pivotal predator (Pycnopodia helianthoides). Sci. Adv. 5, eaau7042 (2019).CAS PubMed PubMed Central Google Scholar
- Hamilton, S. L. et al. Disease-driven mass mortality event leads to widespread extirpation and variable recovery potential of a marine predator across the eastern Pacific. Proc. R. Soc. B 288, 20211195 (2021).CAS PubMed PubMed Central Google Scholar
- Gravem, S. A. et al. Sunflower Sea Star (Pycnopodia helianthoides) (IUCN Red List of Threatened Species, 2021).
- Lowry, D. et al. Endangered Species Act Status Review Report: Sunflower Sea Star (Pycnopodia helianthoides) (National Marine Fisheries Service, Office of Protected Resources, 2022).
- Galloway, A. W. E. et al. Sunflower star predation on urchins can facilitate kelp forest recovery. Proc. R. Soc. B 290, 20221897 (2023).CAS PubMed PubMed Central Google Scholar
- Heady, W. N. et al. Roadmap to Recovery for the Sunflower Sea Star (Pycnopodia helianthoides) along the West Coast of North America (The Nature Conservancy, 2022).
- Hewson, I. et al. Investigating the complex association between viral ecology, environment, and Northeast Pacific sea star wasting. Front. Mar. Sci. 5, 77 (2018).Google Scholar
- Hewson, I., Johnson, M. R. & Reyes-Chavez, B. Lessons learned from the sea star wasting disease investigation. Ann. Rev. Mar. Sci. 17, 2.1–2.23 (2025).Google Scholar
- Jackson, E. W., Pepe-Ranney, C., Johnson, M. R., Distel, D. L. & Hewson, I. A highly prevalent and pervasive densovirus discovered among sea stars from the North American Atlantic Coast. Appl. Environ. Microbiol. 86, e02723-19 (2020).
- Jackson, E. W. et al. Diversity of sea star-associated densoviruses and transcribed endogenous viral elements of densovirus origin. J. Virol. 95, e01594-20 (2021).
- Lloyd, M. M. & Pespeni, M. H. Microbiome shifts with onset and progression of sea star wasting disease revealed through time course sampling. Sci. Rep. 8, 16476 (2018).PubMed PubMed Central Google Scholar
- Aquino, C. A. et al. Evidence that microorganisms at the animal–water interface drive sea star wasting disease. Front. Microbiol. 11, 610009 (2021).PubMed PubMed Central Google Scholar
- Schiebelhut, L. M., DeBiasse, M. B., Gabriel, L., Hoff, K. J. & Dawson, M. N. A reference genome for ecological restoration of the sunflower sea star, Pycnopodia helianthoides. J. Hered. 115, 86–93 (2023).PubMed Central Google Scholar
- Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).CAS PubMed PubMed Central Google Scholar
- Bolyen, E. et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).CAS PubMed PubMed Central Google Scholar
- Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018).PubMed PubMed Central Google Scholar
- McDonald, D. et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 42, 715–718 (2024).CAS PubMed Google Scholar
- Lin, H. & Das Peddada, S. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514 (2020).CAS PubMed PubMed Central Google Scholar
- Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N. & Martinez-Urtaza, J. Non-cholera Vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84 (2017).CAS PubMed Google Scholar
- Gehman, A.-L. M. et al. Fjord oceanographic dynamics provide refuge for critically endangered Pycnopodia helianthoides. Proc. R. Soc. B 292, 20242770 (2025).PubMed PubMed Central Google Scholar
- Menge, B. A. et al. Sea star wasting disease in the keystone predator Pisaster ochraceus in Oregon: insights into differential population impacts, recovery, predation rate, and temperature effects from long-term research. PLoS ONE 11, e0153994 (2016).PubMed PubMed Central Google Scholar
- Suttle, C. A., Chen, F. & Chan, A. M. in International Marine Biotechnology Conference IMBC-91: Short Communications of the Invited Lectures (ed. Nash, C. C.) 153–163 (W. Brown,1992).
- Suttle, C. A. & Chen, F. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58, 3721–3729 (1992).CAS PubMed PubMed Central Google Scholar
- Middleboe, M., Chan, A. M. & Bertelsen, S. K. in Manual of Aquatic Viral Ecology (eds Wilhelm, S. W. et al.) 118–133 (ASLO, 2010).
- Zhong, K. X. et al. Draft genome sequence of Vibrio pectenicida strain FHCF-3, a causative agent of sea star wasting disease in the sunflower sea star (Pycnopodia helianthoides), reveals the genetic potential to produce aerolysin-like toxins. Microbiol. Resour. Announc. (in the press).
- Lambert, C., Nicolas, J. L., Cilia, V. & Corre, S. Vibrio pectenicida sp. nov., a pathogen of scallop (Pecten maximus) larvae. Int. J. Syst. Bacteriol. 48, 481–487 (1998).PubMed Google Scholar
- McCracken, A. R. et al. Microbial dysbiosis precedes signs of sea star wasting disease in wild populations of Pycnopodia helianthoides. Front. Mar. Sci. 10, 1130912 (2023).Google Scholar
- Nicolas, J. L., Corre, S., Gauthier, G., Robert, R. & Ansquer, D. Bacterial problems associated with scallop Pecten maximus larval culture. Dis. Aquat. Organ. 27, 67–76 (1996).Google Scholar
- Lambert, C. & Nicolas, J. L. Specific inhibition of chemiluminescent activity by pathogenic Vibrios in hemocytes of two marine bivalves: Pecten maximus and Crassostrea gigas. J. Invertebr. Pathol. 71, 53–63 (1998).CAS PubMed Google Scholar
- Sandlund, N., Torkildsen, L., Magnesen, T., Mortensen, S. & Bergh, Ø. Immunohistochemistry of great scallop Pecten maximus larvae experimentally challenged with pathogenic bacteria. Dis. Aquat. Organ. 69, 163–173 (2006).PubMed Google Scholar
- Kesarcodi-Watson, A., Miner, P., Nicolas, J. L. & Robert, R. Protective effect of four potential probiotics against pathogen-challenge of the larvae of three bivalves: Pacific oyster (Crassostrea gigas), flat oyster (Ostrea edulis) and scallop (Pecten maximus). Aquaculture 344–349, 29–34 (2012).Google Scholar
- Kesarcodi-Watson, A., Miner, P., Nicolas, J. L., Asmani, K. & Robert, R. Pathogenic threats and probiotic use in larviculture of the scallop, Pecten maximus. Aquac. Res. 47, 1221–1230 (2016).Google Scholar
- Lambert, C., Nicolas, J. L. & Bultel, V. Toxicity to bivalve hemocytes of pathogenic Vibriocytoplasmic extract. J. Invertebr. Pathol. 77, 165–172 (2001).CAS PubMed Google Scholar
- Kehlet-Delgado, H., Häse, C. C. & Mueller, R. S. Comparative genomic analysis of Vibrios yields insights into genes associated with virulence towards C. gigas larvae. BMC Genom.21, 599 (2020).CAS Google Scholar
- GBIF Occurrence Download (GBIF, accessed 5 May 2025); https://doi.org/10.15468/dl.ast4b7
- Kanungo, K. in Invertebrate Blood (ed. Cheng, T. C.) 7–39 (Springer, 1984).
- Hodin, J., Pearson-Lund, A., Anteau, F. P., Kitaeff, P. & Cefalu, S. Progress toward complete life cycle culturing of the endangered sunflower star, Pycnopodia helianthoides. Biol. Bull. 241, 3 (2021).Google Scholar
- Prentice, M. B. et al. Vibrio pectenicida strain FHC F-3 is a causative agent of sea star wasting disease. Dryad https://doi.org/10.5061/dryad.5mkkwh7g9 (2025).
- Montecino-Latorre, D. et al. Devastating transboundary impacts of sea star wasting disease on subtidal asteroids. PLoS ONE 11, e0163190 (2016).PubMed PubMed Central Google Scholar
- Fuess, L. E. et al. Up in arms: immune and nervous system response to sea star wasting disease. PLoS ONE 10, e0133053-16 (2015).
- Brooks, M. E. et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400 (2017).Google Scholar
- Lüdecke, D. sjPlot: Data visualization for statistics in social science. R package version 2.8.17 https://CRAN.R-project.org/package=sjPlot (2024).
- Apprill, A., McNally, S., Parsons, R. & Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137 (2015).Google Scholar
- Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).CAS PubMed PubMed Central Google Scholar
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data (2010); www.bioinformatics.babraham.ac.uk/projects/fastqc
- Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).CAS PubMed PubMed Central Google Scholar
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
- Danecek et al. Twelve years of SAMools and BCFtools. Gigascience 10, gia008 (2021).Google Scholar
- Bushmanova, E., Antipov, D., Lapidus, A. & Prjibelski, A. D. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 8, giz100 (2019).PubMed PubMed Central Google Scholar
- Huson, D. H. et al. MEGAN Community Edition: interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016).PubMed PubMed Central Google Scholar
- R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2024).
- Oksanen, J. et al. Vegan: Community ecology package. R package version 2.7-0 https://github.com/vegandevs/vegan (2024).
- Wickham, H. ggplot2: Elegant graphics for data analysis. R package version 3.5.1 https://ggplot2.tidyverse.org (2016).
- Hester, J. & Bryan, J. glue: Interpreted string literals. R package version 1.8.0 https://glue.tidyverse.org (2024).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011).Google Scholar
- Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).CAS PubMed PubMed Central Google Scholar
- Davis, N. M., Proctor, D., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).PubMed PubMed Central Google Scholar
- Wick, R. Porechop: adapter trimmer for Oxford Nanopore reads, version 0.2. GitHubhttps://github.com/rrwick/Porechop (2018).
- Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).PubMed PubMed Central Google Scholar
- Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).CAS PubMed PubMed Central Google Scholar
- Dong, X. & Strous, M. An integrated pipeline for annotation and visualization of metagenomic contigs. Front. Genet. 10, 999 (2019).CAS PubMed PubMed Central Google Scholar
- Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).CAS PubMed Google Scholar
- Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and useability. Mol. Biol. Evol. 30, 772–780 (2013).CAS PubMed PubMed Central Google Scholar
- Steenwyk, J. L., Buida, T. J. III, Li, Y., Shen, X.-X. & Rokas, A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenetic inference. PLoS Biol. 18, e3001007 (2020).CAS PubMed PubMed Central Google Scholar
- Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274 (2015).CAS PubMed Google Scholar
- Yoon, S.-H., Ha, S.-M., Lim, J. M., Kwon, S. J. & Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton. Leeuw. 110, 1281–1286 (2017).CAS Google Scholar
